|
Cancer Systems and Integrative Biology
by Usha N. Kasid (Editor), Robert Clarke (Editor) BOOK CHAPTER 22, DOI: https://doi.org/10.1007/978-1-0716-3163-8_22, 2023. Single Cell and Spatial Analysis of Emergent Organoid Platforms [Link] Aditi Kumar, Shuangyi Cai, Mayar Allam, Samuel Henderson, Melissa Ozbeyler, Lilly Saiontz, and Ahmet F. Coskun This chapter presents growth strategies, molecular screening methods, and emerging issues of organoid platforms. Single-cell and spatial analysis resolve organoid heterogeneity to obtain information about the structural and molecular cellular states. An essential resource is an organoid atlas that can catalog protocols and standardize data analysis for different organoid types. Molecular profiling of individual cells in organoids and data organization of the organoid landscape will impact biomedical applications from basic science to translational use. |
|
Revealing Uncharted Biology with Single Cell Multiplex Proteomic Technologies
by Editor: Wendy Fantl BOOK CHAPTER 8, eBook ISBN: 9780128222102. High-definition CODEX for 3D multiplex spatial cell phenotyping [Link] Thomas Hu, Nicholas Zhang, Mythreye Venkatesan, Christian M. Schürch, Garry P. Nolan, and Ahmet F. Coskun We present a high-definition CODEX (HD-CODEX)that integrates a spinning disk confocal imaging with conventional CODEX chemistry, yielding multiplexed proteomic images of whole cells in 3D at sub-100-nm resolution. In this superresolved CODEX data, deep learning-based cell segmentation reconstructed individual cell phenotypes and molecular neighborhoods in lymph nodes and nasopharyngeal tissues. |
Spatial immunophenotyping using multiplexed imaging of immune follicles in secondary lymphoid tissues
|
|
PREDICTIVE SPATIAL MORPHO-PROTEOMICS |
Spatial Morphoproteomic Features Predict Uniqueness of Immune Microarchitectures and Responses in Lymphoid Follicles
BiorXiv, doi: https://doi.org/10.1101/2024.01.05.574186 (2024) [Link]
Thomas Hu , Mayar Allam, Vikram Kaushik, Steven L Goudy, Qin Xu, Pamela Mudd, Kalpana Manthiram, and Ahmet F Coskun
Multiplex imaging technologies allow the characterization of single cells in their cellular environments. Understanding the organization of single cells within their microenvironment and quantifying disease-status related biomarkers is essential for multiplex datasets. Here we proposed SNOWFLAKE, a graph neural network framework pipeline for the prediction of disease-status from combined multiplex cell expression and morphology in human B-cell follicles. We applied SNOWFLAKE to a multiplex dataset related to COVID-19 infection in humans and showed better predictive power of the SNOWFLAKE pipeline compared to other machine learning and deep learning methods. Moreover, we combined morphological features inside graph edge features to utilize attribution methods for extracting disease-relevant motifs from single-cell spatial graphs. The underlying subgraphs were further analyzed and associated with disease status across the dataset. We showed that SNOWFLAKE successfully extracted significant low dimensional embedding from subgraphs with a clear separation between disease status and helped characterize unique cellular interactions in the subgraphs. SNOWFLAKE is a generalizable pipeline for the analysis of multiplex imaging data modality by extracting disease-relevant subgraphs guided by graph-level prediction.
|
SINGLE CELL SPATIAL METABOLOMICS |
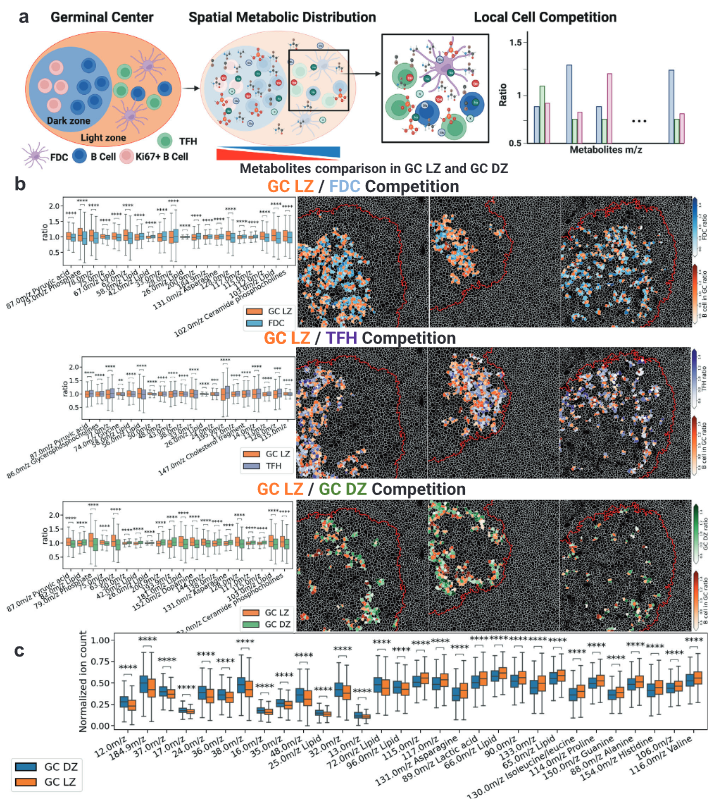
Single-cell spatial metabolomics with cell-type specific protein profiling for tissue systems biology
Nature Communications, volume 14, Article number: 8260 (2023) [Link]
Thomas Hu , Mayar Allam, Shuangyi Cai , Walter Henderson , Brian Yueh , Aybuke Garipcan , Anton Ievlev , Maryam Afkarian , Semir Beyaz, and Ahmet F Coskun
Metabolic reprogramming in cancer and immune cells occurs to support their increasing energy needs in biological tissues. Here we propose Single Cell Spatially resolved Metabolic (scSpaMet) framework for joint protein-metabolite profiling of single immune and cancer cells in male human tissues by incorporating untargeted spatial metabolomics and targeted multiplexed protein imaging in a single pipeline. We utilized the scSpaMet to profile cell types and spatial metabolomic maps of 19507, 31156, and 8215 single cells in human lung cancer, tonsil, and endometrium tissues, respectively. The scSpaMet analysis revealed cell type-dependent metabolite profiles and local metabolite competition of neighboring single cells in human tissues. Deep learning-based joint embedding revealed unique metabolite states within cell types. Trajectory inference showed metabolic patterns along cell differentiation paths. Here we show scSpaMet’s ability to quantify and visualize the cell-type specific and spatially resolved metabolic-protein mapping as an emerging tool for systems-level understanding of tissue biology.
|
SPATIAL TRANSCRIPTOMICS - ORGANOIDS |
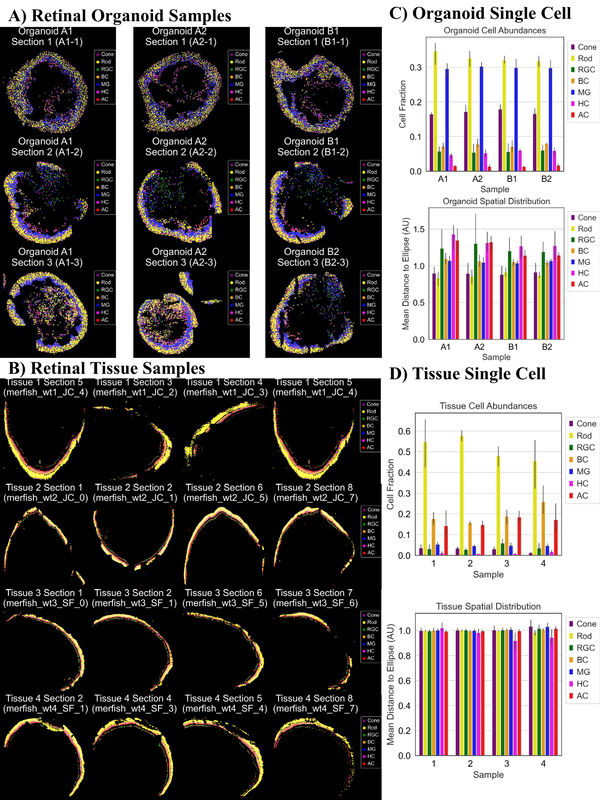
Tissue Spatial Omics Dissects Organoid Biomimicry
GEN Biotechnology, Volume 2, Number 5 DOI: 10.1089/genbio.2023.0039 (2023) [Link]
Nicholas Zhang, Denis Ohlstrom, Sicheng Pang, Nivik Sanjay Bharadwaj, Aaron Qu, Hans Grossniklaus, and Ahmet F. Coskun
Recently, organoids, or three-dimensional (3D) cellular assemblies, have demonstrated promise as viable models for organ development and disease study. In contrast to challenging preclinical models, organoids are advantageous due to rapid fabrication times and greater patient specificity. The advent of spatial transcriptomics and single cell technologies has also enhanced the characterization of intraorganoid heterogeneity, thus highlighting 3D cell signaling and organ development at micro scales. In this study, we describe ongoing and future directions in spatial omics integrated with various imaging technologies for two-dimensional/3D organoid characterization. Utilizing both retinal organoids and native retinal tissues, we undertook an analysis to deconstruct the cellular compositions and structural attributes of their respective cell layers. Our findings indicate that the spatial organization of cell phenotypes is similar between organoids and native retinal tissue. However, it is noteworthy that native retinal tissue possesses thinner yet distinctly separated cell layers compared with the organoids.
|
SPATIAL OMICS - VOICES |
What do you most hope spatial molecular profiling will help us understand? Part 2: "Enabling time travel and future telling in hierarchical tissue maps"
Cell Systems (2023) [Link]
Itai Yanai, Elana J. Fertig, Mingyao Li, Fabian Coscia, Johanna Klughammer, Qing Nie, Jinmiao Chen, and Ahmet F. Coskun
|
SUBCELLULAR SPATIAL TRANSCRIPTOMICS |
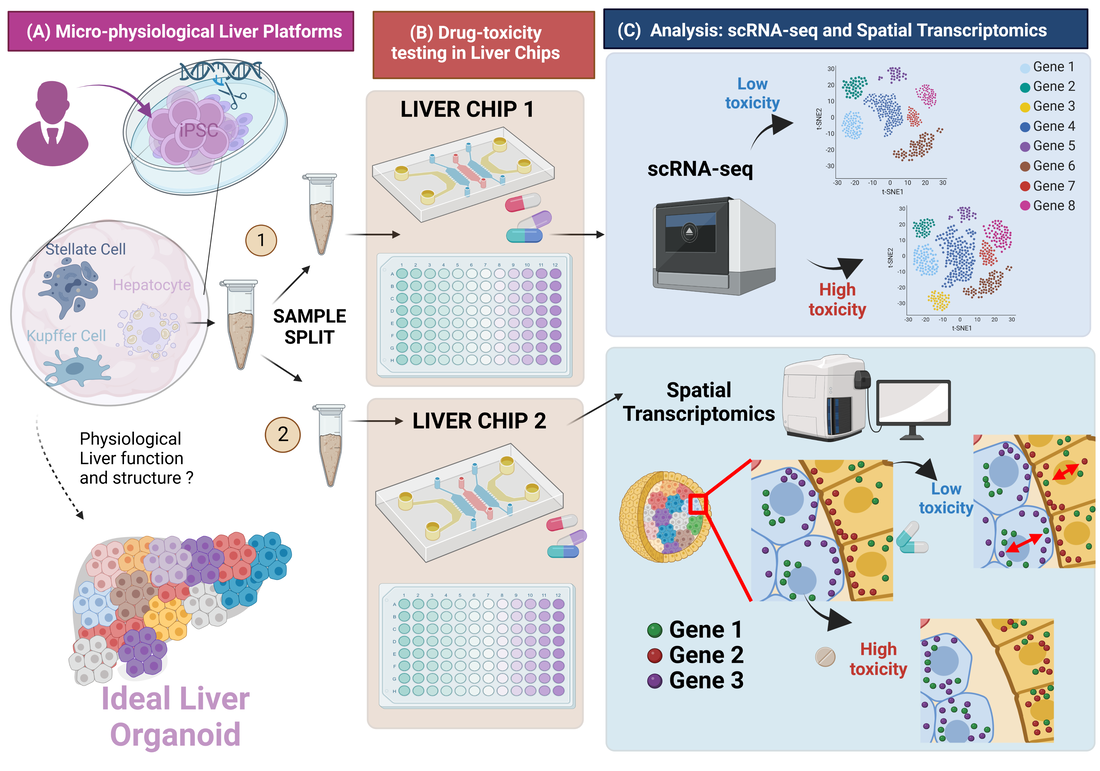
Single-cell RNA-seq and subcellular spatial transcriptomics facilitate the translation of liver microphysiological systems for regulatory application
Journal of Pharmaceutical Analysis (2023) [Link]
Dan Li, Zhou Fang, Qiang Shi, Nicholas Zhang, Binsheng Gong, Weida Tong, Ahmet F. Coskun, Joshua Xu.
scRNA-seq and spatial transcriptomics have become increasingly valuable tools in understanding cellular heterogeneity and organization in a wide range of contexts, including in vitro models and disease states. However, these technologies also present significant challenges, particularly in terms of quality assurance. In addition, MPS is still in its infancy and the technology is rapid evolving. It is unlikely that MPS can completely replace animal tests in the near future. Despite these challenges, the continued development and improvement of quality control standards
and guidelines for single-cell sequencing and spatial transcriptomics are necessary to ensure reliable and reproducible results in future research. With the growing interest and investment in these fields, there are numerous opportunities for advancements in technology and data analysis, as well as exciting prospects for new discoveries and therapeutic interventions.
and guidelines for single-cell sequencing and spatial transcriptomics are necessary to ensure reliable and reproducible results in future research. With the growing interest and investment in these fields, there are numerous opportunities for advancements in technology and data analysis, as well as exciting prospects for new discoveries and therapeutic interventions.
|
SUBCELLULAR SPATIAL TRANSCRIPTOMICS |
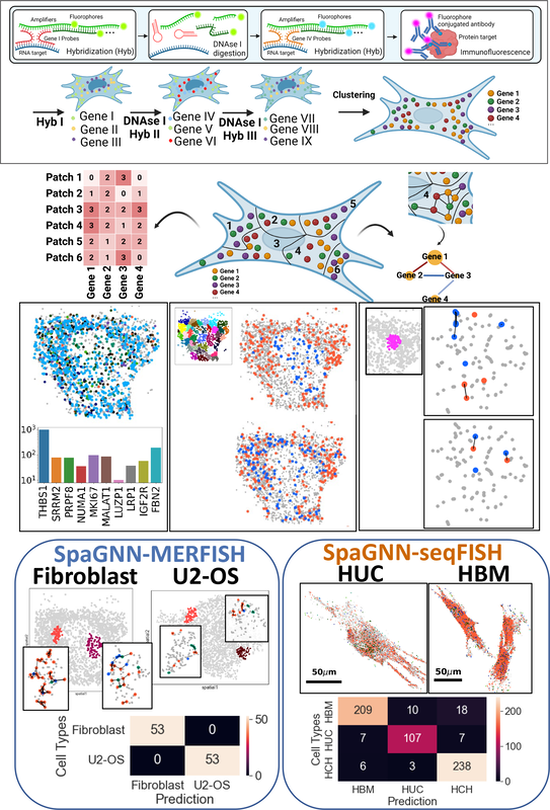
Subcellular spatially resolved gene neighborhood networks in single cells
Cell Reports Methods, 100476 (2023) [Link]
Zhou Fang, Adam Ford, Thomas Hu, Nicholas Zhang, Athanasios Mantalaris, and Ahmet F. Coskun
Image-based spatial omics methods such as fluorescence in situ hybridization (FISH) generate molecular profiles of single cells at single-molecule resolution. Current spatial transcriptomics methods focus on the distribution of single genes. However, the spatial proximity of RNA transcripts can play an important role in cellular function. We demonstrate a spatially resolved gene neighborhood network (spaGNN) pipeline for the analysis of subcellular gene proximity relationships. In spaGNN, machine-learning-based clustering of subcellular spatial transcriptomics data yields subcellular density classes of multiplexed transcript features. The nearest-neighbor analysis produces heterogeneous gene proximity maps in distinct subcellular regions. We illustrate the cell-type-distinguishing capability of spaGNN using multiplexed error-robust FISH data of fibroblast and U2-OS cells and sequential FISH data of mesenchymal stem cells (MSCs), revealing tissue-source-specific MSC transcriptomics and spatial distribution characteristics. Overall, the spaGNN approach expands the spatial features that can be used for cell-type classification tasks.
News article: A Look Inside Stem Cells Helps Create Personalized Regenerative Medicine
News article: A Look Inside Stem Cells Helps Create Personalized Regenerative Medicine
Spatial subcellular organelle networks in single cells
|
Combinatorial treatment rescues tumour-microenvironment-mediated attenuation of MALT1 inhibitors in B-cell lymphomas
Nature Materials (2023) [Link]
Shivem B. Shah, Christopher R. Carlson, Kristine Lai, Zhe Zhong, Grazia Marsico, Katherine M. Lee, Nicole E. Félix Vélez, Elisabeth B. Abeles, Mayar Allam, Thomas Hu, Lauren D. Walter, Karen E. Martin, Khanjan Gandhi, Scott D. Butler, Rishi Puri, Angela L. McCleary-Wheeler, Wayne Tam, Olivier Elemento, Katsuyoshi Takata, Christian Steidl, David W. Scott, Lorena Fontan, Hideki Ueno, Benjamin D. Cosgrove, Giorgio Inghirami, Andrés J. García, Ahmet F. Coskun, Jean L. Koff, Ari Melnick & Ankur Singh
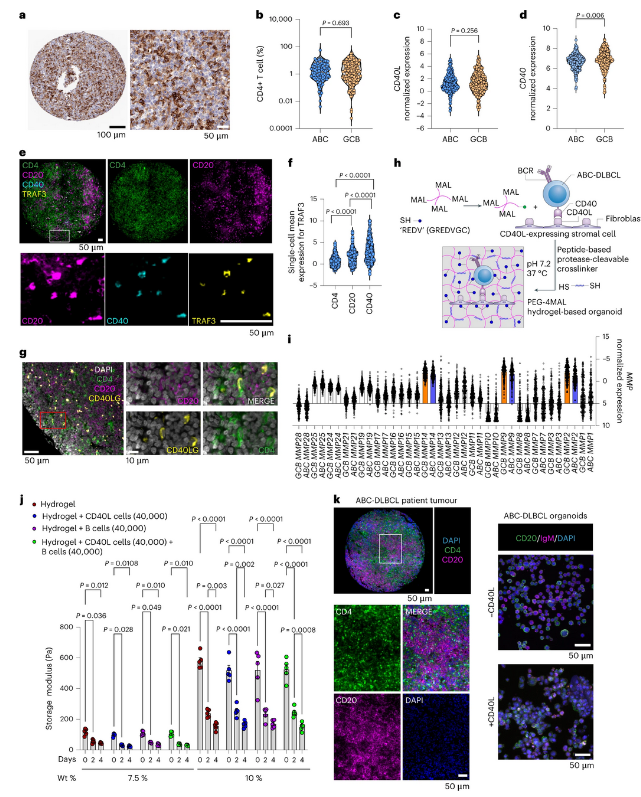
Activated B-cell-like diffuse large B-cell lymphomas (ABC-DLBCLs) are characterized by constitutive activation of nuclear factor κB driven by the B-cell receptor (BCR) and Toll-like receptor (TLR) pathways. However, BCR-pathway-targeted therapies have limited impact on DLBCLs. Here we used >1,100 DLBCL patient samples to determine immune and extracellular matrix cues in the lymphoid tumour microenvironment (Ly-TME) and built representative synthetic-hydrogel-based B-cell-lymphoma organoids accordingly. We demonstrate that Ly-TME cellular and biophysical factors amplify the BCR–MYD88–TLR9 multiprotein supercomplex and induce cooperative signalling pathways in ABC-DLBCL cells, which reduce the efficacy of compounds targeting the BCR pathway members Bruton tyrosine kinase and mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1). Combinatorial inhibition of multiple aberrant signalling pathways induced higher antitumour efficacy in lymphoid organoids and implanted ABC-DLBCL patient tumours in vivo. Our studies define the complex crosstalk between malignant ABC-DLBCL cells and Ly-TME, and provide rational combinatorial therapies that rescue Ly-TME-mediated attenuation of treatment response to MALT1 inhibitors.
|
SPATIAL PROTEOMICS - PRECISION ONCOLOGY |
Spatially variant immune infiltration scoring in human cancer tissues
Npj Precision Oncology, 6, Article number: 60 (2022) [Link]
Mayar Allam ,Thomas Hu ,Jeongjin Lee , Jeffrey Aldrich , Sunil Badve , Yesim Polar , Manali Bhave , Suresh Ramalingam , Frank Schneider, and Ahmet F. Coskun
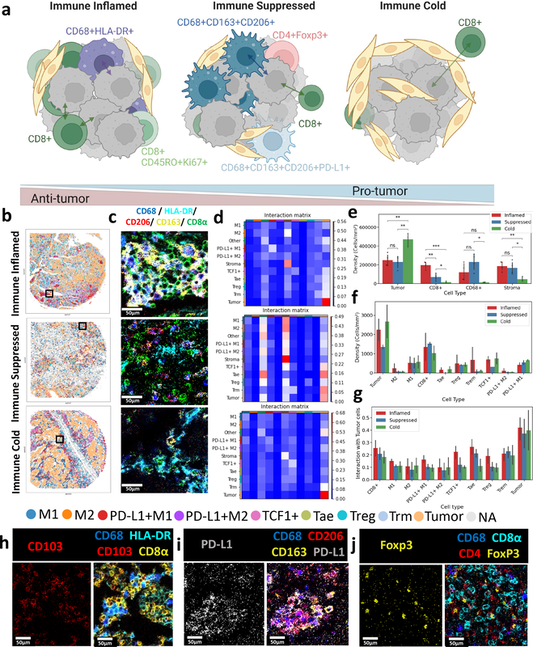
The Immunoscore is a method to quantify the immune cell infiltration within cancers to predict the disease prognosis. Previous immune profiling approaches relied on limited immune markers to establish patients’ tumor immunity. However, immune cells exhibit a higher-level complexity that is typically not obtained by the conventional immunohistochemistry methods. Herein, we present a spatially variant immune infiltration score, termed as SpatialVizScore, to quantify immune cells infiltration within lung tumor samples using multiplex protein imaging data. Imaging mass cytometry (IMC) was used to target 26 markers in tumors to identify stromal, immune, and cancer cell states within 26 human tissues from lung cancer patients. Unsupervised clustering methods dissected the spatial infiltration of cells in tissue using the high-dimensional analysis of 16 immune markers and other cancer and stroma enriched labels to profile alterations in the tumors’ immune infiltration patterns. Spatially resolved maps of distinct tumors determined the spatial proximity and neighborhoods of immune-cancer cell pairs. These SpatialVizScore maps provided a ranking of patients’ tumors consisting of immune inflamed, immune suppressed, and immune cold states, demonstrating the tumor’s immune continuum assigned to three distinct infiltration score ranges. Several inflammatory and suppressive immune markers were used to establish the cell-based scoring schemes at the single-cell and pixel-level, depicting the cellular spectra in diverse lung tissues. Thus, SpatialVizScore is an emerging quantitative method to deeply study tumor immunology in cancer tissues.
|
SUBCELLULAR SPATIAL PROTEOMICS |
Multiplexed protein profiling reveals spatial subcellular signaling networks
iScience, DOI:10.1016/j.isci.2022.104980 (2022) [Link]
Shuangyi Cai, Thomas Hu, Mythreye Venkatesan, Mayar Allam, Suresh Ramalingam, Shi-Yong Sun, and Ahmet F. Coskun
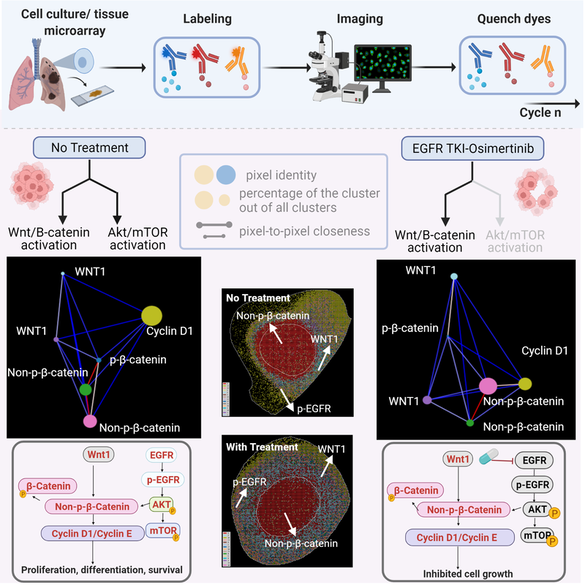
Protein-protein interaction networks are altered in multi-gene dysregulations in many disorders. Image-based protein multiplexing sheds light on signaling pathways to dissect cell-to-cell heterogeneity, previously masked by the bulk assays. Herein, a rapid multiplexed immunofluorescence (RapMIF) method measured up to 25-plex spatial protein maps from cultures and tissues at subcellular resolution, providing combinatorial 272 pair-wise and 1360 tri-protein signaling states across 33 multiplexed pixel-level clusters. The RapMIF pipeline automated staining, bleaching, and imaging of the biospecimens in a single platform. The RapMIF data revealed the compensatory role of spatial signaling networks for AKT/mTOR and WNT/β-catenin pathways. Subcellular protein images demonstrated translocation patterns, spatial receptor discontinuity, and subcellular signaling clusters in single cancer cells. Signaling networks exhibited spatial redistribution of signaling proteins in drug-responsive cultures. Machine learning analysis predicted the phosphorylated β-catenin expression from interconnected signaling protein images. RapMIF is an ideal signaling discovery approach for precision therapy design.
|
SPATIAL OMICS OF ORGANOIDS |
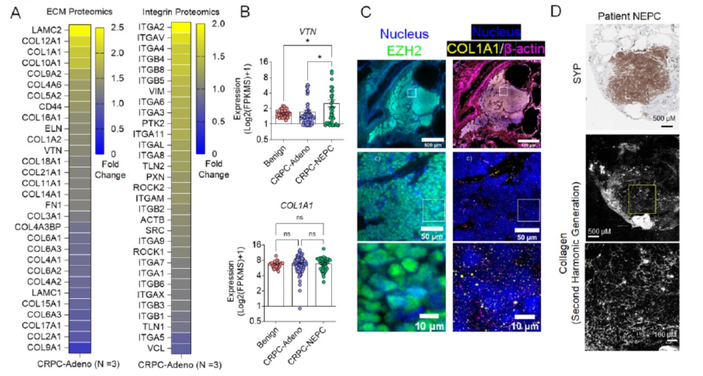
Extracellular Matrix in Synthetic Hydrogel-based Prostate Cancer Organoids Regulate Therapeutic Response to EZH2 and DRD2 inhibitors,
Advanced Materials (2022) [Link]
Matthew J Mosquera, Sungwoong Kim, Rohan Bareja, Zhou Fang, Shuangyi Cai, Heng Pan, Muhammad Asad, M. Laura Martin, Michael Sigouros, Florencia M Rowdo, Sarah Ackermann, Jared Capuano, Jacob Bernheim, Cynthia Cheung, Ashley Doane, Nicholas Brady, Richa Singh, David S. Rickman, Varun Prabhu, Joshua E Allen, Loredana Puca, Ahmet F Coskun, Mark Rubin, Himisha Beltran, Juan Miguel Mosquera, Olivier Elemento, Ankur Singh
Following treatment with androgen receptor (AR) pathway inhibitors, ∼20% of prostate cancer patients progress by shedding their AR dependence. These tumors undergo epigenetic reprogramming turning castration-resistant prostate cancer adenocarcinoma (CRPC-Adeno) into neuroendocrine prostate cancer (CRPC-NEPC). Currently, no targeted therapies are available for CRPC-NEPCs, and there are minimal organoid models to discover new therapeutic targets against these aggressive tumors. Here, using a combination of patient tumor proteomics, RNA sequencing, spatial omics, immunohistochemistry, and a synthetic hydrogel-based organoid, we define putative extracellular matrix (ECM) cues that regulate the phenotypic, transcriptomic, and epigenetic underpinnings of CRPC-NEPCs. Short-term culture in tumor-expressed ECM differentially regulated DNA methylation and mobilized genes in CRPC-NEPC tumors. The ECM type distinctly regulated the response to small molecule inhibitors of epigenetic repressor EZH2 and Dopamine Receptor D2 (DRD2), the latter being an understudied target in neuroendocrine tumors. In vivo patient-derived xenograft studies in immunocompromised mice showed a robust anti-tumor response when treated with a DRD2 inhibitor. Finally, we demonstrate that therapeutic response in CRPC-NEPCs under drug-resistant ECM conditions can be overcome by first cellular reprogramming with EZH2 inhibitors, followed by DRD2 treatment. The synthetic hydrogel-based organoids suggest the regulatory role ECM may play in therapeutic response to targeted therapies in CRPC-NEPCs and enable the discovery of single drugs and combination therapies to overcome resistance.
|
SPATIAL PROTEOMICS |
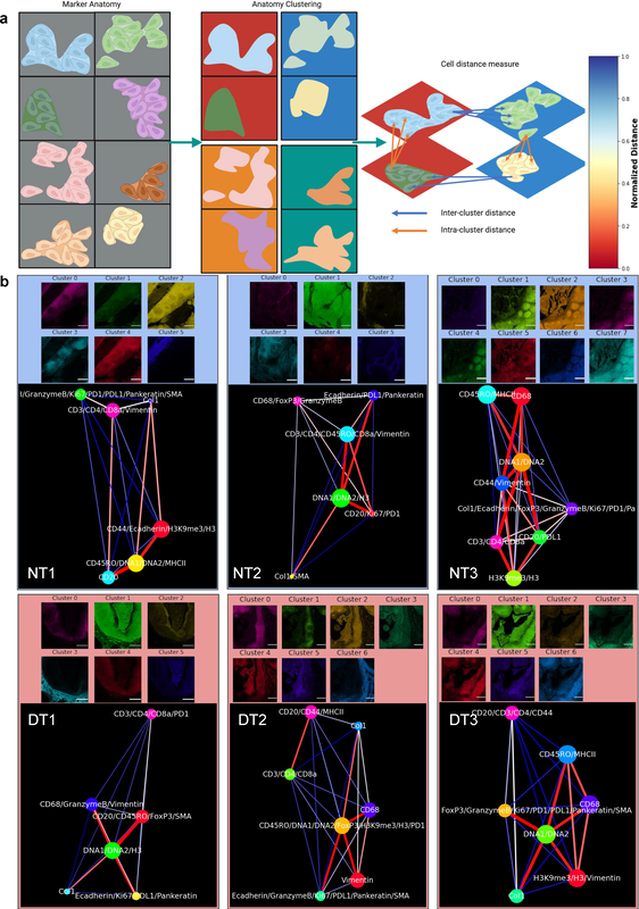
Spatially visualized single-cell pathology of highly multiplexed protein profiles in health and disease
Communications Biology, 4, 632 (2021) [Link]
Mayar Allam, Thomas Hu, Shuangyi Cai, Krishman Laxminarayanan, Robert B. Hughley, and Ahmet F. Coskun
Deep molecular profiling of biological tissues is an indicator of health and disease. As one of the mainstream technologies, we used imaging mass cytometry (IMC) to acquire 20-plex spatially resolved, multiparameter protein data in tissue sections from normal and chronic tonsillitis cases. To quantify this high-dimensional IMC data, herein we present SpatialViz, a suite of algorithms to explore spatial relationships in multiplexed tissue images using the nexus of single-cell quantifications as a bottom-up method and computational pathology of anatomical distributions as a top-down strategy. Single-cell and spatial maps confirmed that CD68+ cells were correlated with the enhanced Granzyme B expression and CD3+ cells exhibited enrichment of CD4+ phenotype in chronic tonsillitis. SpatialViz revealed morphological distributions of cellular organizations in distinct anatomical areas and spatially resolved associations of single-cells by intra-cluster and the inter-cluster analysis across anatomical categories. Spatial proximity analysis showed distance maps between the marker-pairs. Spatial topographic maps showed the distinct organization of tissue layers with unique multiplex marker profiles. The spatial reference framework generated network-based comparisons of multiplex data from health and diseased tonsils. SpatialViz is a general pipeline to visualize and quantify single-cell granularity and anatomical complexity in diverse multiplexed tissue imaging data.
|
SPATIAL METABOLOMICS |
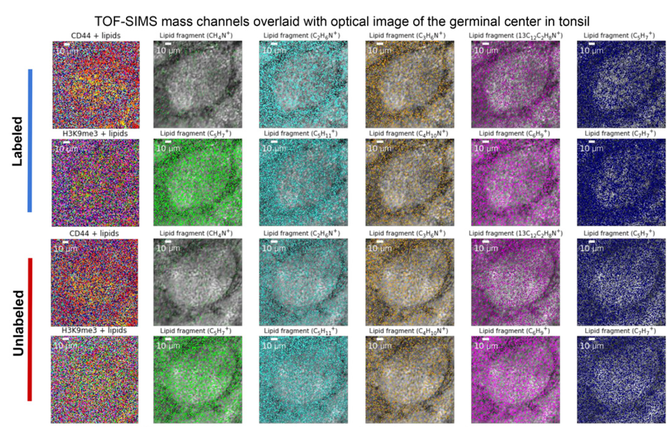
Spatially resolved 3D metabolomic profiling in tissues
Science Advances, Vol. 7, no. 5, eabd0957 (2021) [Link]
Shambavi Ganesh, Thomas Hu, Eric Woods, Mayar Allam, Shuangyi Cai, Walter Henderson, and Ahmet F. Coskun
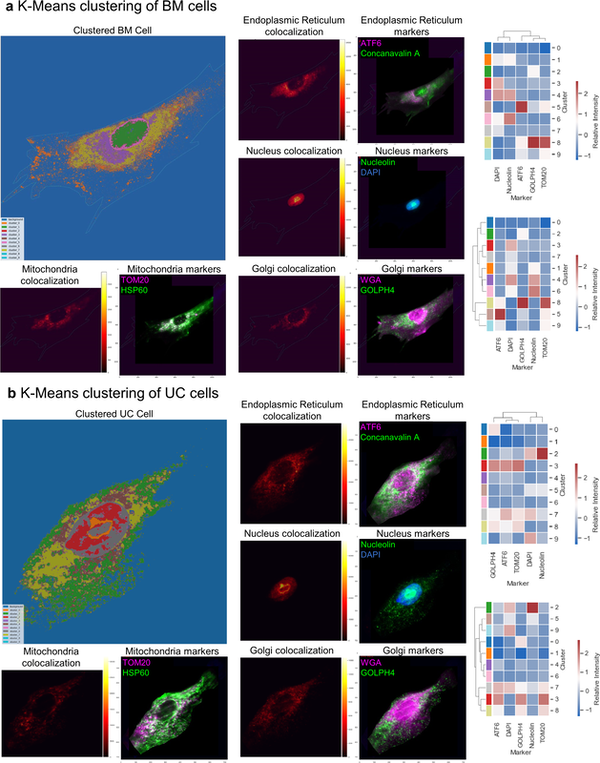
Spatially-resolved RNA and protein molecular analyses have revealed surprising heterogeneity of cells. Metabolic analysis of individual cells has the potential to complement these single-cell studies. Herein, we present a 3D Spatially-resolved Metabolomics Framework, 3D-SMF, to map out the spatial organization of metabolites and cell-features in immune cells of human tonsils. In this method, 3D metabolic profiles were acquired by a Time-of-Flight Secondary Ion Mass Spectrometry (TOF-SIMS) instrument to profile up to 188 compounds. Ion beams were used to measure sub-10-nm layers of tissue through z-stacks with up to 150 sections of a tonsil. To incorporate cell-specificity, tonsil tissues were labeled by an isotope-tagged antibody library. To explore relations of metabolic and cellular features, data reduction, 3D spatial correlations, unsupervised K-means clustering, and network analyses were carried out. Immune cells exhibited spatially distinct lipidomics distributions in lymphatic tissue. The 3D-SMF pipeline impacts studying the immune cells in health and disease.
|
SINGLE CELL ANALYSIS |
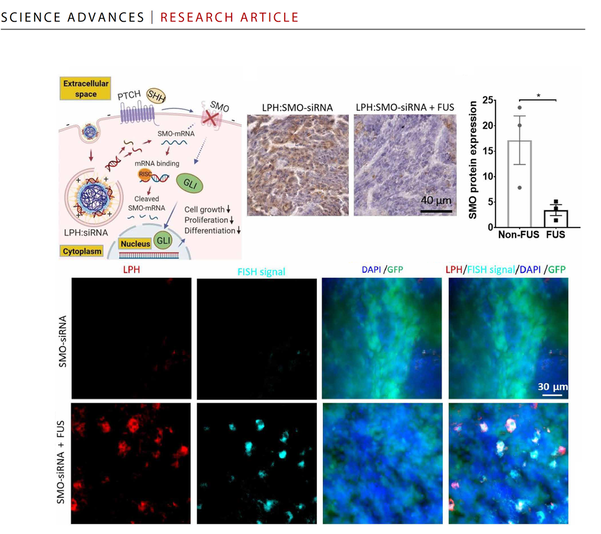
Single-cell analysis reveals effective siRNA delivery in brain tumors with microbubble-enhanced ultrasound and cationic nanoparticles
Science Advances (2021) DOI: 10.1126/sciadv.abf7390 [Link]
Yutong Guo, Hohyun Lee, Zhou Fang, Anastasia Velalopoulou, Jinhwan Kim, Midhun Ben Thomas, Jingbo Liu, Ryan G. Abramowitz, YongTae Kim, Ahmet F. Coskun, Daniel Pomeranz Krummel, Soma Sengupta, Tobey J. MacDonald and Costas Arvanitis
RNA-based therapies offer unique advantages for treating brain tumors. However, tumor penetrance and uptake are hampered by RNA therapeutic size, charge, and need to be “packaged” in large carriers to improve bioavailability. Here, we have examined delivery of siRNA, packaged in 50-nm cationic lipid-polymer hybrid nanoparticles (LPHs:siRNA), combined with microbubble-enhanced focused ultrasound (MB-FUS) in pediatric and adult preclinical brain tumor models. Using single-cell image analysis, we show that MB-FUS in combination with LPHs:siRNA leads to more than 10-fold improvement in siRNA delivery into brain tumor microenvironments of the two models. MB-FUS delivery of Smoothened (SMO) targeting siRNAs reduces SMO protein production and markedly increases tumor cell death in the SMO-activated medulloblastoma model. Moreover, our analysis reveals that MB-FUS and nanoparticle properties can be optimized to maximize delivery in the brain tumor microenvironment, thereby serving as a platform for developing next-generation tunable delivery systems for RNA-based therapy in brain tumors.
|
SUBCELLULAR SPATIAL OMICS |
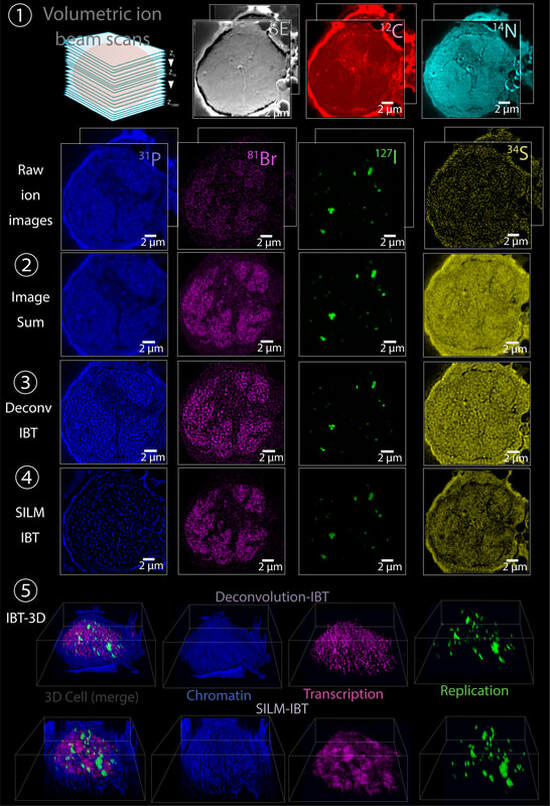
Nanoscopic subcellular imaging enabled by Ion Beam Tomography
Nature Communications, 12, Article number: 789 (2021) [Link]
Ahmet F. Coskun, Guojun Han, Shambavi Ganesh, Shih-Yu Chen , Xavier Rovira-Clave, Stefan Harmsen, Sizun Jiang , Christian Schürch, Yunhao Bai , Chuck Hitzman and Garry P. Nolan
Multiplexed ion beam imaging (MIBI) has been previously used to profile multiple parameters in two dimensions in single cells within tissue slices. Here, a mathematical and technical framework for three-dimensional (3D) subcellular MIBI is presented. Ion beam tomography (IBT) compiles ion beam images that are acquired iteratively across successive, multiple scans, and later assembled into a 3D format without loss of depth resolution. Algorithmic deconvolution, tailored for ion beams, is then applied to the transformed ion image series, yielding 4-fold enhanced ion beam data cubes. To further generate 3D sub-ion-beam width precision visuals, isolated ion molecules are localized in the raw ion beam images, creating an approach coined as SILM, secondary ion beam localization microscopy, providing sub-25-nm accuracy in original ion images. Using deep learning, a parameter-free reconstruction method for ion beam tomograms with high accuracy is developed for low-density targets. In cultured cancer cells and tissues, IBT enables accessible visualization of 3D volumetric distributions of genomic regions, RNA transcripts, and protein factors with 5-nm axial resolution using isotope-enrichments and label-free elemental analysis. Multiparameter imaging of subcellular features at near macromolecular resolution is implemented by the IBT tools as a general biocomputation pipeline for imaging mass spectrometry.
|
SPATIAL PROTEOMICS AND PHARMACO-IMAGING |
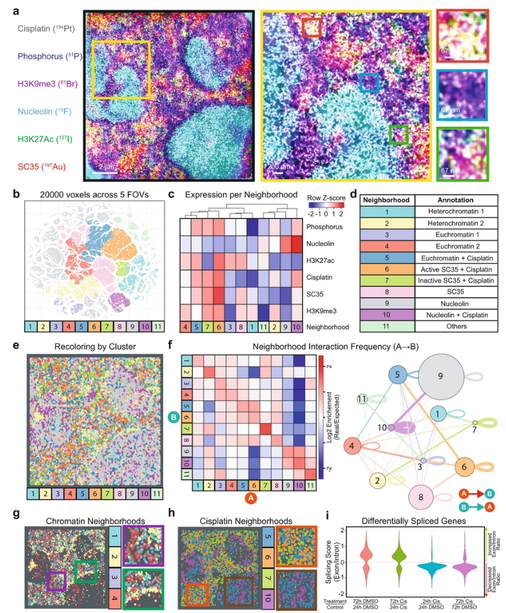
Subcellular localization of biomolecules and drug distribution by high-definition ion beam imaging
Nature Communications, 12, 4628 (2021).
Simultaneous visualization of the relationship between multiple biomolecules and their ligands or small molecules at the nanometer scale in cells will enable greater understanding of how biological processes operate. We present here high-definition multiplex ion beam imaging (HD-MIBI), a secondary ion mass spectrometry approach capable of high-parameter imaging in 3D of targeted biological entities and exogenously added structurally-unmodified small molecules. With this technology, the atomic constituents of the biomolecules themselves can be used in our system as the “tag” and we demonstrate measurements down to ~30 nm lateral resolution. We correlated the subcellular localization of the chemotherapy drug cisplatin simultaneously with five subnuclear structures. Cisplatin was preferentially enriched in nuclear speckles and excluded from closed-chromatin regions, indicative of a role for cisplatin in active regions of chromatin. Unexpectedly, cells surviving multi-drug treatment with cisplatin and the BET inhibitor JQ1 demonstrated near total cisplatin exclusion from the nucleus, suggesting that selective subcellular drug relocalization may modulate resistance to this important chemotherapeutic treatment. Multiplexed high-resolution imaging techniques, such as HD-MIBI, will enable studies of biomolecules and drug distributions in biologically relevant subcellular microenvironments by visualizing the processes themselves in concert, rather than inferring mechanism through surrogate analyses.
|
BIOE MEDIA LAB |
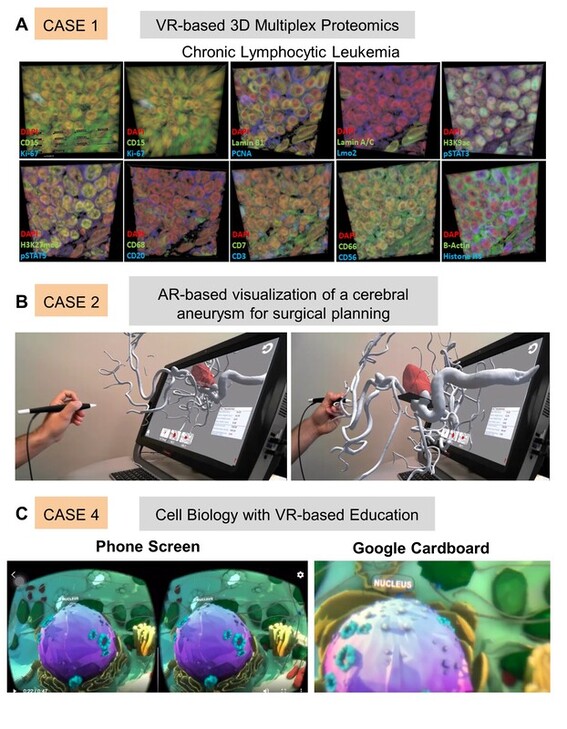
Virtual and Augmented Reality for Biomedical Applications
Cell Reports Medicine, DOI: 10.1016/j.xcrm.2021.100348 (2021) [Link]
Mythreye Venkatesan, Harini Mohan, Justin Ryan, Christian Schuerch, Garry Nolan, David Frakes, and Ahmet F. Coskun
3D visualization technologies such as virtual reality (VR), augmented reality (AR), and mixed reality (MR) have gained popularity in the recent decade. Digital extended reality (XR) technologies have been adopted in
various domains ranging from entertainment to education because of their accessibility and affordability. XR modalities create an immersive experience, enabling 3D visualization of the content without a conventional 2D display constraint. Here, we provide a perspective on XR in current biomedical applications and demonstrate case studies using cell biology concepts, multiplexed proteomics images, surgical data for heart operations, and cardiac 3D models. Emerging challenges associated with XR technologies in the context of adverse health effects and a cost comparison of distinct platforms are discussed. The presented XR platforms will be useful for biomedical education, medical training, surgical guidance, and molecular data visualization to enhance trainees’ and students’ learning, medical operation accuracy, and the comprehensibility of complex biological systems.
various domains ranging from entertainment to education because of their accessibility and affordability. XR modalities create an immersive experience, enabling 3D visualization of the content without a conventional 2D display constraint. Here, we provide a perspective on XR in current biomedical applications and demonstrate case studies using cell biology concepts, multiplexed proteomics images, surgical data for heart operations, and cardiac 3D models. Emerging challenges associated with XR technologies in the context of adverse health effects and a cost comparison of distinct platforms are discussed. The presented XR platforms will be useful for biomedical education, medical training, surgical guidance, and molecular data visualization to enhance trainees’ and students’ learning, medical operation accuracy, and the comprehensibility of complex biological systems.
|
PUBLIC HEALTH |
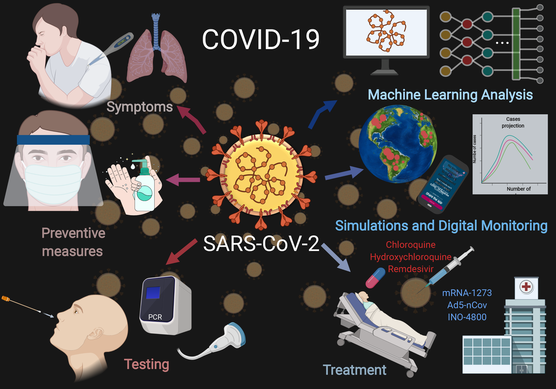
COVID-19 Diagnostics, Tools, and Prevention
Diagnostics (2020) [PDF] Special Issue "Virus Diagnostic Methods: Learning from the COVID-19 Global Outbreak"
Mayar Allam, Shuangyi Cai, Shambavi Ganesh, Mythreye Venkatesan , Saurabh Doodhwala, Zexing Song, Thomas Hu Kaiwen, Aditi Kumar, Jeremy Heit, Kasfia Kazi, Pushti Desai, Shivam Patel, Harini Mohan, Melissa Ozbeyler, Samuel Henderson, Andrew Borst, Beliz Utebay, and Ahmet F. Coskun
The Coronavirus Disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), outbreak from Wuhan City, Hubei province, China in 2019 has become an ongoing global health emergency. The emerging virus, SARS-CoV-2, causes coughing, fever, muscle ache, and shortness of breath or dyspnea in symptomatic patients. The pathogenic particles that are generated by coughing and sneezing remain suspended in the air or attach to a surface to facilitate transmission in an aerosol form. This review focuses on the recent trends in pandemic biology, diagnostics methods, prevention tools, and policies for COVID-19 management. To meet the growing demand for medical supplies during the COVID-19 era, a variety of personal protective equipment (PPE) and ventilators have been developed using do-it-yourself (DIY) manufacturing. COVID-19 diagnosis and the prediction of virus transmission are analyzed by machine learning algorithms, simulations, and digital monitoring. Until the discovery of a clinically approved vaccine for COVID-19, pandemics remain a public concern. Therefore, technological developments, biomedical research, and policy development are needed to decipher the coronavirus mechanism and epidemiological characteristics, prevent transmission, and develop therapeutic drugs.
|
SPATIAL BIOLOGY |
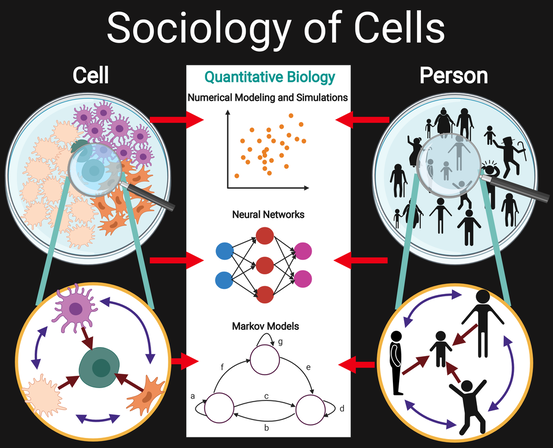
Cellular sociology regulates the hierarchical spatial patterning and organization of cells in organisms
Open Biology, (2020) [PDF]
Shambavi Ganesh*, Beliz Utebay*, Jeremy Heit* & Ahmet F. Coskun
Advances in biotechnology have increasingly revealed interactions of cells with their surroundings, suggesting a cellular society at the microscale. Similarities between cells and humans across multiple hierarchical levels have inference potential for reaching quantitative insights. Here, the functional and structural comparisons between how cells and individuals fundamentally socialize to give rise to spatial organization are hypothesized. Integrative experimental and computational methods, coupled with emerging biochemical methodologies, shape the societal perspective of cellular interactions that create spatially coordinated forms in biological systems. Emerging quantitative models from simpler biological microworld and single cell organisms are investigated, providing a route to model spatio-temporal regulation of humans. This analogical reasoning framework sheds light on patterning principles of biological sociology from the cellular scale and up.
|
SPATIAL PRECISION ONCOLOGY |
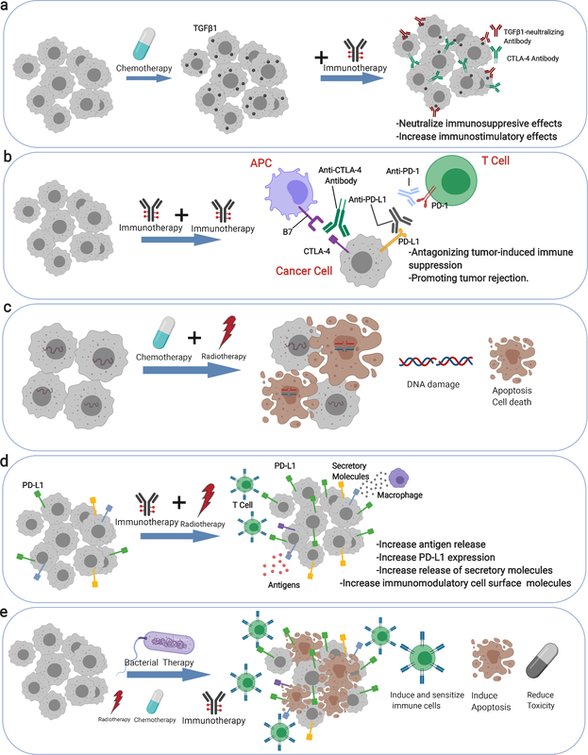
Multiplex Spatial Bioimaging for Combination Therapy Design
Trends in Cancer (2020) [PDF]
Shuangyi Cai*, Mayar Allam* & Ahmet F. Coskun
This paper presents combination therapies to address the current gap in the field of cancer therapies. Combination therapy administers synergistic drugs to patients for cancer treatments. However, a subset of patients remains unresponsive due to tumor heterogeneity. Combination therapy overcomes this problem to eliminate cancers with reduced off-targets, resistance, and relapse. Multiplex bioimaging aids in determining the molecular mechanisms in patient biopsies, informing the design of multi-drug therapies.
|
SPATIAL PRECISION ONCOLOGY |
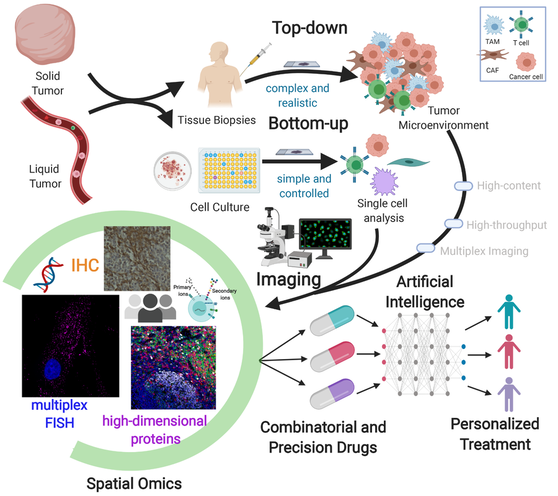
Multiplex bioimaging of single-cell spatial profiles for precision cancer diagnostics and therapeutics
NPJ Precision Oncology, vol 4, 11 (2020) [PDF]
Mayar Allam*, Shuangyi Cai* & Ahmet F. Coskun
Nature Research Cancer Community | Behind the Paper Story
Cancers exhibit functional and structural diversity in distinct patients. In this mass, normal and malignant cells create tumor microenvironment that is heterogeneous among patients. A residue from primary tumors leaks into the bloodstream as cell clusters and single cells, providing clues about disease progression and therapeutic response. The complexity of these hierarchical microenvironments needs to be elucidated. While tumors comprise ample cell-types, the standard clinical technique is still the histology that is limited to a single marker. Multiplexed imaging technologies open new directions in pathology. Spatially resolved proteomic, genomic, and metabolic profiles of human cancers are now possible at the single-cell level. This perspective discusses spatial bioimaging methods to decipher the cascade of microenvironments in solid and liquid biopsies. A unique synthesis of top-down and bottom-up analysis methods is presented. Spatial multi-omics profiles can be tailored to precision oncology through artificial intelligence. Data-driven patient profiling enables personalized medicine and beyond.
Cancers exhibit functional and structural diversity in distinct patients. In this mass, normal and malignant cells create tumor microenvironment that is heterogeneous among patients. A residue from primary tumors leaks into the bloodstream as cell clusters and single cells, providing clues about disease progression and therapeutic response. The complexity of these hierarchical microenvironments needs to be elucidated. While tumors comprise ample cell-types, the standard clinical technique is still the histology that is limited to a single marker. Multiplexed imaging technologies open new directions in pathology. Spatially resolved proteomic, genomic, and metabolic profiles of human cancers are now possible at the single-cell level. This perspective discusses spatial bioimaging methods to decipher the cascade of microenvironments in solid and liquid biopsies. A unique synthesis of top-down and bottom-up analysis methods is presented. Spatial multi-omics profiles can be tailored to precision oncology through artificial intelligence. Data-driven patient profiling enables personalized medicine and beyond.
|
BIOE MEDIA LAB |
Digital posters for interactive cellular media and bioengineering education
Communications Biology, 2, 455 (2019) [PDF]
Mythreye Venkatesan & Ahmet F. Coskun
Georgia Tech BME News | Petit Institute News
Conventional posters are effective in disseminating progress reports in scientific meetings, but they fail to deliver the need for visualization of dynamic biological data and become costly with the increasing number of conferences and the reprinting needs for emerging research. Here we present digital posters that repurpose digital frames from the art community and experiment with multiplexed imaging movies of cells as a demonstration of the digital poster concept, providing an interactive and low-cost tool for next-generation sharing platforms.
Conventional posters are effective in disseminating progress reports in scientific meetings, but they fail to deliver the need for visualization of dynamic biological data and become costly with the increasing number of conferences and the reprinting needs for emerging research. Here we present digital posters that repurpose digital frames from the art community and experiment with multiplexed imaging movies of cells as a demonstration of the digital poster concept, providing an interactive and low-cost tool for next-generation sharing platforms.
|
BARCODED BIOIMAGING |
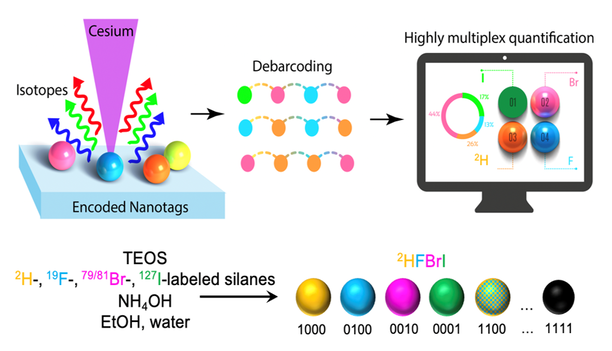
Isotopically encoded nanotags for multiplexed ion beam imaging
Advanced Materials Technology, 2000098 (2020) [PDF]
Stefan Harmsen*, Ahmet F. Coskun*, Shambavi Ganesh, Garry P. Nolan and Sam Gambhir
High-dimensional profiling of markers and analytes using approaches such as barcoded fluorescent imaging with repeated labeling and mass cytometry has allowed visualization of biological processes at the single-cell level. To address limitations of sensitivity and mass-channel capacity, we have developed a nano-barcoding platform for multiplexed ion beam imaging (MIBI) using secondary ion beam spectrometry that utilizes fabricated isotopically encoded nanotags. Use of combinatorial isotope distributions in 100-nm-sized nanotags expanded the labeling palette to overcome the spectral bounds of mass channels. As a proof-of-principle, a four-digit (i.e., 0001 to 1111) barcoding scheme was demonstrated to detect 16 variants of 2H, 15N, 19F, 79/81Br and 127I elemental barcode sets that were encoded in silica nanoparticle matrices. A computational debarcoding method and an automated machine learning analysis approach were developed to extract barcodes for accurate quantification of spatial nanotag distributions in large ion beam imaging areas up to 0.6 mm2. Isotopically encoded nanotags should boost the performance of mass imaging platforms such as MIBI and other elemental-based bioimaging approaches.
|
SPATIAL GENOMICS |
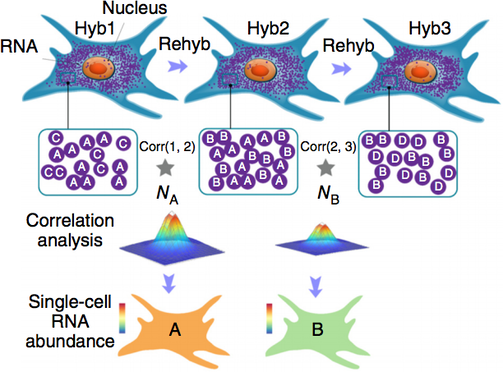
Dense transcript profiling in single cells by image correlation decoding,
Nature Methods 2016
Sequential barcoded fluorescent in situ hybridization (seqFISH) allows large numbers of molecular species to be accurately detected in single cells, but multiplexing is limited by the density of barcoded objects. We present correlation FISH (corrFISH), a method to resolve dense temporal barcodes in sequential hybridization experiments. Using corrFISH, we quantified highly expressed ribosomal protein genes in single cultured cells and mouse thymus sections, revealing cell-type-specific gene expression.
|
SPATIAL GENOMICS |
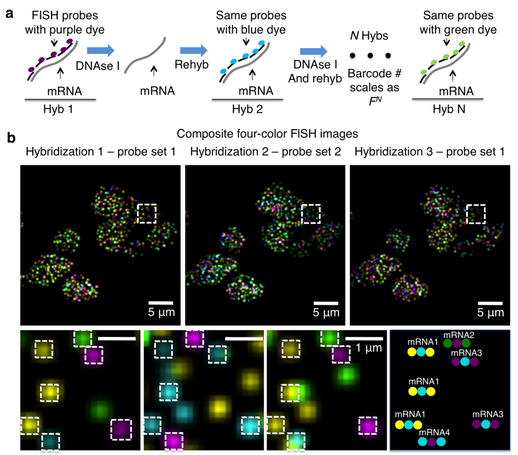
Single-cell in situ RNA profiling by sequential hybridization,
Nature Methods 2014
In this Correspondence, we present a sequential barcoding scheme to multiplex different mRNAs.Here, the mRNAs in cells are barcoded by sequential rounds of hybridization, imaging and probe stripping. As the transcripts are fixed in cells, the corresponding fluorescent spots remain in place during multiple rounds of hybridization and can be aligned to read out a fluorophore sequence. This sequential barcode is designed to uniquely identify an mRNA
|
BIOIMAGING |
Wide-field optical detection of nanoparticles using on-chip microscopy and self-assembled nanolenses,
Nature Photonics 2013
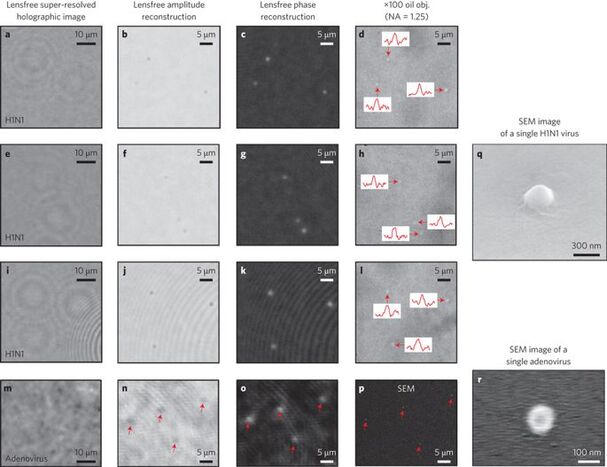
The direct observation of nanoscale objects is a challenging task for optical microscopy because the scattering from an individual nanoparticle is typically weak at optical wavelengths. Electron microscopy therefore remains one of the gold standard visualization methods for nanoparticles, despite its high cost, limited throughput and restricted field-of-view. Here, we describe a high-throughput, on-chip detection scheme that uses biocompatible wetting films to self-assemble aspheric liquid nanolenses around individual nanoparticles to enhance the contrast between the scattered and background light. We model the effect of the nanolens as a spatial phase mask centred on the particle and show that the holographic diffraction pattern of this effective phase mask allows detection of sub-100 nm particles across a large field-of-view of >20 mm2. As a proof-of-concept demonstration, we report on-chip detection of individual polystyrene nanoparticles, adenoviruses and influenza A (H1N1) viral particles.
|
BIOIMAGING |
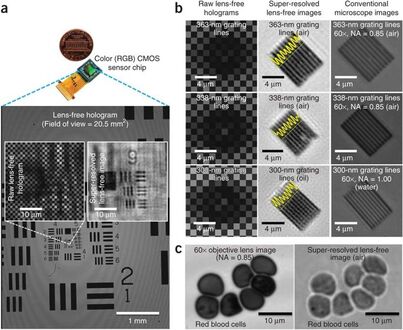
Imaging without lenses: achievements and remaining challenges of wide-field on-chip microscopy,
Nature Methods 2012
We discuss unique features of lens-free computational imaging tools and report some of their emerging results for wide-field on-chip microscopy, such as the achievement of a numerical aperture (NA) of ∼0.8–0.9 across a field of view (FOV) of more than 20 mm2 or an NA of ∼0.1 across a FOV of ∼18 cm2, which corresponds to an image with more than 1.5 gigapixels. We also discuss the current challenges that these computational on-chip microscopes face, shedding light on their future directions and applications.